Advanced R for Econometricians
Functional Style R Programming
Martin C. Arnold, Jens Klenke
Part I
Functional Programming Using purrr
Functional Style Programming
To become significantly more reliable, code must become more transparent. In particular, nested conditions and loops must be viewed with great suspicion. Complicated control flows confuse programmers. Messy code often hides bugs. — Bjarne Stroustrup
FAQs
# Packages neededlibrary(dplyr)library(purrr)library(readr)# (or simply attach the tidyverse!)What is a functional programming (FP) language?
Simply put: a language which is centered on problem-solving using functions!
There are two common threads in FP:
A functional language has first-class functions which behave like any other data type. In R this means we may treat function like variables (i.e., assign them, store them in a
listor pass them as arguments to other functions)Many functional languages require pure functions only. These are functions which have no side-effects: they do not interfere with anything outside their scope and produce output which depends only on the input.
R has first-class functions.
Does R allow for pure functions only? No: e.g.
print()has side-effects. Obviously all functions which return pseudo-random numbers are not pure functions.
FAQs
Is R a FP language?
R is not a FP language but we may adopt a functional style of programming.
Functionals are functions which take functions as input and produce, e.g. vector output
Why should I use functional style programming?
FP is often space efficient, very comprehensible and easily adopted to new situations
Functionals are easily analysed in isolation and thus are often straightforward to optimise and parallelise
(We'll discuss functionals in a minute)
You've probably used functionals already: lapply() and integrate() are prominent examples.
FAQs
What does 'FP style' even mean?
It’s hard to describe exactly what a functional style is, but generally we will refer to the following definition:
Functional programming style means decomposing a big problem into smaller pieces, then solving each piece with a function or combination of functions. — Hadley Wickham
FAQs
We will briefly discuss key techniques in functional R programming which are best summarised by the table below. We will focus on purrr functionals and applications of function factories.
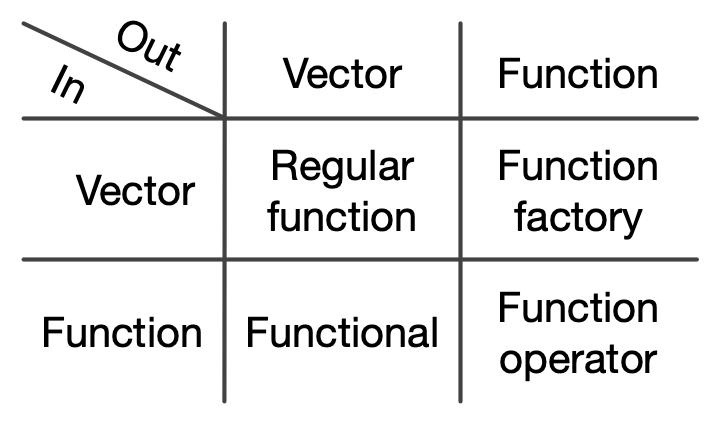
Source: Wickham (2019)
Functionals
A functional takes a function as an input and returns a vector as output.
Let's do something that might seem strange at first sight:
randomise <- function(f) f(rnorm(1e3))randomise(mean)## [1] -0.002711432randomise(sum)## [1] -75.23842Functionals can also produce other data structures, e.g., data frames as output.
Functionals
Functionals are often used as alternatives to loops. Not because loops are inherently slow (which is common wisdom), but because loops
make it relatively cumbersome to harness the power of iteration
are prone to typos that are difficult to identify
can be overly flexible: loops convey that an iteration is done, but may make it relatively hard to grasp what is done and what should be done with the results.
Functionals are tailored to specific tasks which immediately convey why they are being used and what output format they produce.
Switching from loops to functionals doesn't necessarily mean that we must write our own functionals: the
purrrpackage provides functionals which are very easy to apply and also fast as they are written in C.
With functionals we don't need to worry about indexing, brackets, curly braces etc.
Of course, flexibility isn't bad. They idea of a FP is to use functionals that perform a specific iteration which returns a specific output format.
Others are likely to be puzzled by looking at your code if it uses a lot of loops. Functionals immediately convey which iteration is done and which output is returned.
'Others' also includes your future self :-)
Functionals — purrr::map()
map() is the purrr version of lapply().
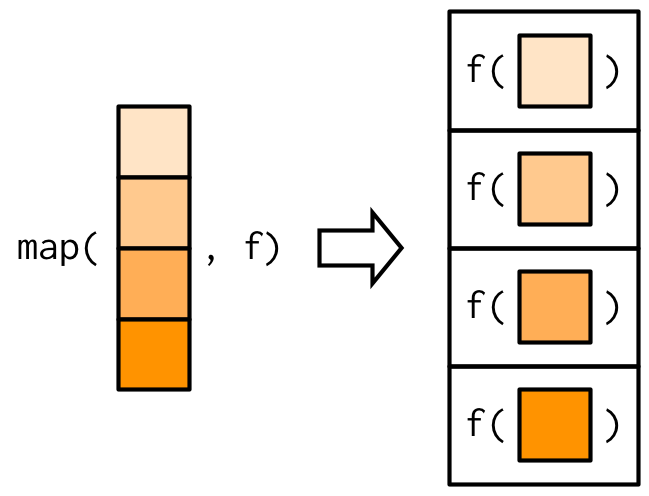
Source: Wickham (2019)
Example: map()
map(1:3, f) is list(f(1), f(2), f(3)).
triple <- function(x) x * 3map(1:3, triple)## [[1]]## [1] 3## ## [[2]]## [1] 6## ## [[3]]## [1] 9Do the example using
lapply():lapply(1:3, function(x) x * 3)Obviously
map()returns a list, too.
purrr::map_*() — Producing Atomic Vectors
There are helper functions which are more convenient if simpler data structures are required:
map_lgl(),map_int(),map_dbl(), andmap_chr()return an atomic vector of the specified typeBase R equivalents are
sapply()andvapply()
Example: map_*()
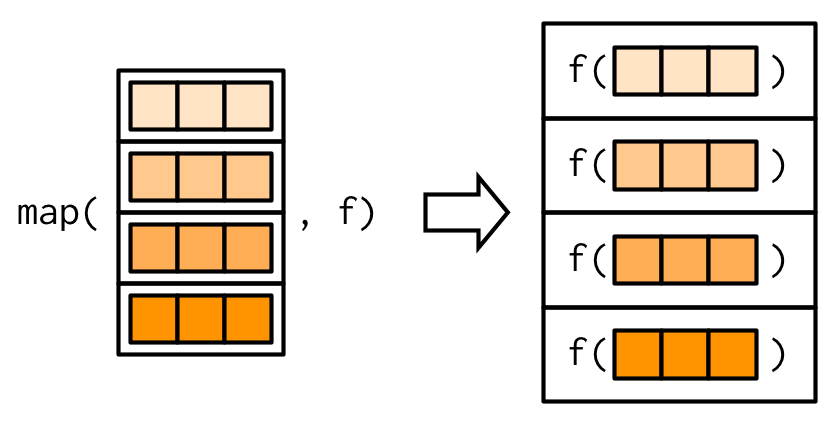
Source: Wickham (2019)
# check class of mtcars dataclass(mtcars)## [1] "data.frame"map_lgl(mtcars, is.double)## mpg cyl disp hp drat wt qsec vs am gear carb ## TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEn_unique <- function(x) length(unique(x))map_int(mtcars, n_unique)## mpg cyl disp hp drat wt qsec vs am gear carb ## 25 3 27 22 22 29 30 2 2 3 6Remember that
logical,integer,doubleandcharacterare atomic typesmtcarsis a data frame thus themap_*()functions map the columnsOf course, the
*inmap_*()must match the return type of the functions used for mapping
purrr::map_*() — Producing Atomic Vectors
The twiddle operator
~allows to write anonymous functions in a less verbose manner. It conveys that the subsequent expression is a formula.A good rule of thumb: if a function spans lines or uses
{...}, we should give it a name.
Example: map_*() with inline anonymous function
map_dbl(mtcars, function(x) length(unique(x)))## mpg cyl disp hp drat wt qsec vs am gear carb ## 25 3 27 22 22 29 30 2 2 3 6map_dbl(mtcars, ~ length(unique(.x)))## mpg cyl disp hp drat wt qsec vs am gear carb ## 25 3 27 22 22 29 30 2 2 3 6purrr::map_*() — Producing Atomic Vectors
map_*() is useful for selecting elements from a list by name, position, or both.
Example: element extraction with map_*()
x <- list( list(-1, x = 1, y = c(2), z = "a"), list(-2, x = 4, y = c(5, 6), z = "b"), list(-3, x = 8, y = c(9, 10, 11)))map_dbl(x, "x") # select by name## [1] 1 4 8map_dbl(x, 1) # select by position## [1] -1 -2 -3In base R we'd have to write a function that iterates through x or use sapply().
purrr::map_*() — Producing Atomic Vectors
Task:
Write short Base R code which works on x from the previous slide and returns
all entries named
'x'andall entries at position 1 from this nested list.
# 1.sapply(x, "[[", "x")# 2.sapply(x, "[[", 1)## [1] 1 4 8## [1] -1 -2 -3purrr::map_*() — Producing Atomic Vectors
Note that components must exist in all entries of the object we iterate over (here a nested list).
Example: element extraction with map_*()
map_chr(x, "z") # z doesn't exist in x[[3]]## Error in `stop_bad_type()`:## ! Result 3 must be a single string, not NULL of length 0To prevent this error a .default value can be supplied.
map_chr(x, "z", .default = NA)## [1] "a" "b" NAKeep in mind that this requires your subsequent code to work with NAs.
purrr::map_*() — Producing Atomic Vectors
Additional arguments to the mapping function may be passed after the function name.
We need to be careful with evaluation!
Example: mapping with additional arguments
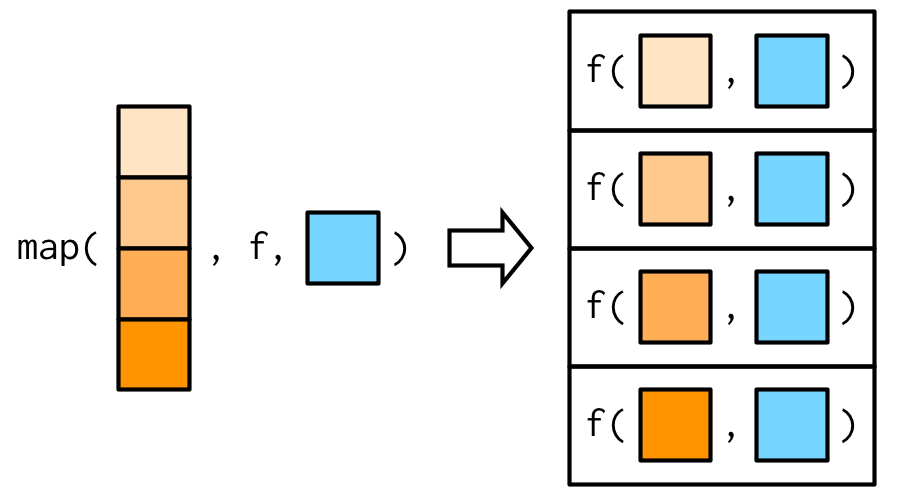
Source: Wickham (2019)
x <- list(1:5, c(1:10, NA))map_dbl(x, ~ mean(.x, na.rm = TRUE))## [1] 3.0 5.5More efficient:
map_dbl(x, mean, na.rm = TRUE)## [1] 3.0 5.5Arguments passed to an anonymous function are evaluated in every iteration. The latter approach is more efficient because additional arguments are evaluated just once.
purrr::map_*() — Producing Atomic Vectors
Additional arguments are not decomposed: map_*() is only vectorised over (the data passed as) the first argument. Further (vector) arguments are passed along.
Example: mapping with additional arguments — ctd.
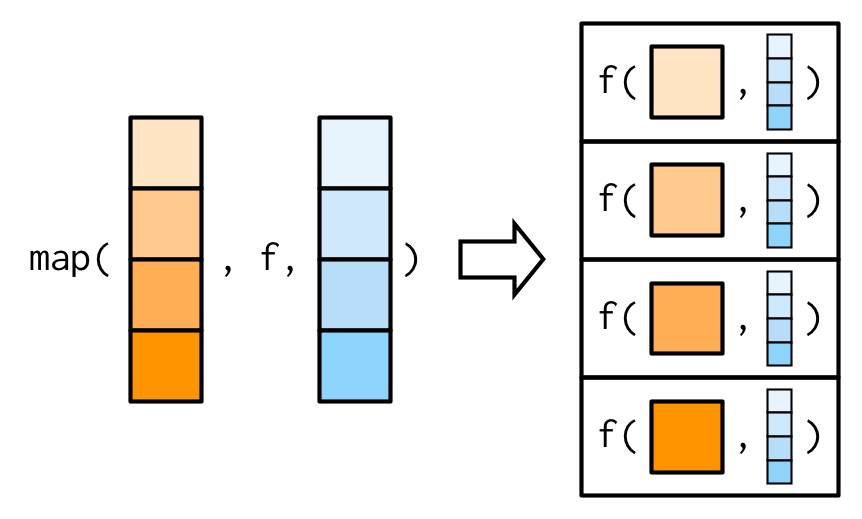
Source: Wickham (2019)
# Arg. 'mean' is recycledmap(1:3, rnorm, mean = c(100, 10, 1))## [[1]]## [1] 101.1215## ## [[2]]## [1] 101.324174 9.246503## ## [[3]]## [1] 101.8168982 10.5856532 0.9970042Question to students:
Explain the outcomes of map(1:4, rnorm, mean = c(100, 10, 1))
purrr::map_*() — Producing Atomic Vectors
Example: mapping over a different argument
Assume you'd like to investigate the impact of different amounts of trimming when computing the mean of observations sampled from a heavy-tailed distribution.
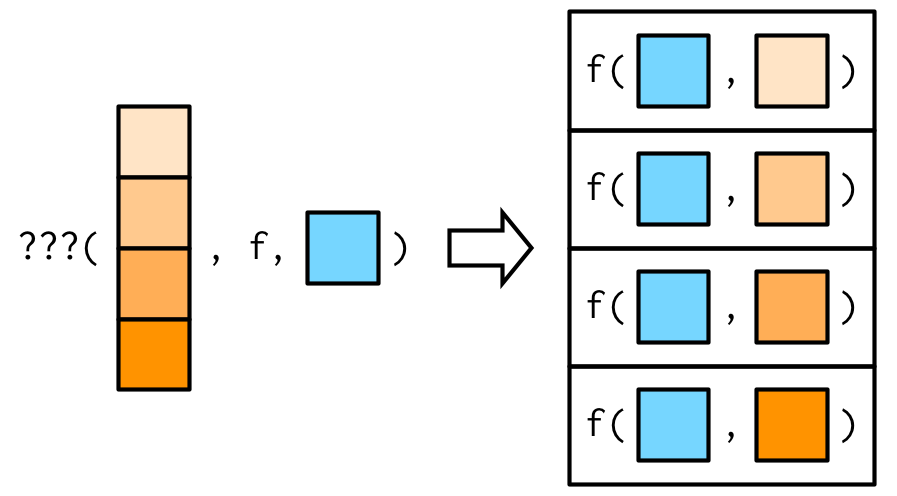
Source: Wickham (2019)
trims <- c(0, 0.1, 0.2, 0.5)x <- rcauchy(1000)We may switch arguments using an anonymous function:
map_dbl(trims, ~ mean(x, trim = .x))## [1] 0.624460050 0.006129521 0.038663335 0.075093275This is equivalent to:
map_dbl(trims, function(trim) mean(x, trim = trim))## [1] 0.624460050 0.006129521 0.038663335 0.075093275purrr::map_*() — Exercises
map(1:3, ~ runif(2))is a useful pattern for generating random numbers, butmap(1:3, runif(2))is not. Why not? Can you explain why it returns the result that it does?The following code simulates the performance of a t-test for non-normal data. Extract the p-value from each test, then visualise.
trials <- map(1:100, ~ t.test(rpois(10, 10), rpois(10, 7)))Use
map()to fit linear models to themtcarsdataset using the formulas stored in this list:formulas <- list(mpg ~ disp,mpg ~ disp + wt,mpg ~ I(1 / disp) + wt)
map(1:3, ~ runif(2))evaluatesrunif()withn = 2in every iteration since~converts to an anonymous function.map(1:3, runif(2))evaluatesrunif(2)only once and cannot do mapping becauserunif(2)is not treated as a function.NULLis returned in every iteration.Code:
library(ggplot2)trials_df <- tibble(p_value = map_dbl(trials, "p.value"))trials_df %>%ggplot(aes(x = p_value, fill = p_value < 0.05)) +geom_histogram(binwidth = .025) +ggtitle("Distribution of p-values for random Poisson data.")Code:
models <- map(formulas, lm, data = mtcars)
Case Study: Model Fitting with purrr
Tired of mtcars? We're too... let's use cars2018, a dataset on fuel efficiency of real cars of today from a US Department of Energy instead! 🚗🚗🚗
We will now take a look at how purrr functions can be used to fit a regression model to subgroups of data, extract estimates and then compare the approach to base R approaches.
Instructions
Load the
cars2018.csvdataset and split it byDrive, see?splitUse
purrr(preferably together withdplyr) to- fit the model
MPG ~ Cylindersto each subgroup - extract the estimated coefficient of
Cylinders
- fit the model
Contrast your
purrrapproach to base R alternatives that rely on*apply()andfor(), respectively
Map Variants
There are 23 variants of map*() which are easily understood as variants of the following functions:
Output same type as input:
modify()Iterate over two inputs:
map2()Iterate with an index:
imap()Return nothing:
walk()Iterate over any number of inputs:
pmap()
| List | Atomic | Same type | Nothing | |
|---|---|---|---|---|
| One argument | map() |
map_*() |
modify() |
walk() |
| Two arguments | map2() |
map2_*() |
modify2() |
walk2() |
| One argument + index | imap() |
imap_*() |
imodify() |
iwalk() |
| N arguments | pmap() |
pmap_*() |
— |
pwalk() |
The table shows input (rows) and output types (columns).
purrr::modify()
The modify() function works on the input components and returns an object of the same type as the input.
Example: data.frame in / data.frame out
df <- data.frame( x = 1:3, y = 6:4)modify(df, ~ .x * 2)## x y## 1 2 12## 2 4 10## 3 6 8Note that modify() never modifies in-place but creates a copy, which must be (re)assigned.
df <- modify(df, ~ .x * 2)purrr::map2()
map2() is vectorised over two arguments.
Example: weighted mean using map2()
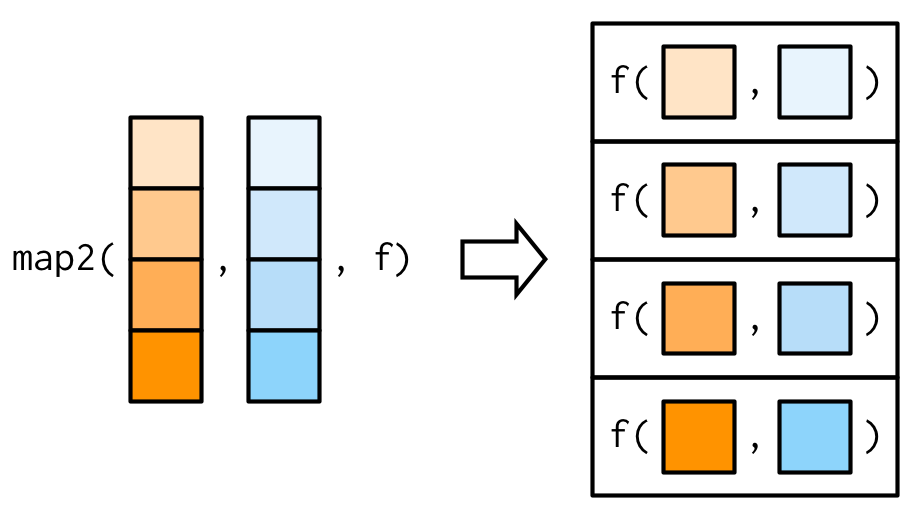
Source: Wickham (2019)
Let's generate lists of observations and associated weights.
set.seed(123)xs <- map(1:4, ~ runif(4))xs[[1]][[1]] <- NAws <- map(1:4, ~ rpois(4, 5) + 1)map2_dbl varies both xs and ws as inputs to weighted.mean().
map2_dbl(xs, ws, weighted.mean)## [1] NA 0.6625391 0.5968213 0.5287878purrr::map2()
Additional arguments may be passed just as with map().
Example: weighted mean using map2() — ctd.
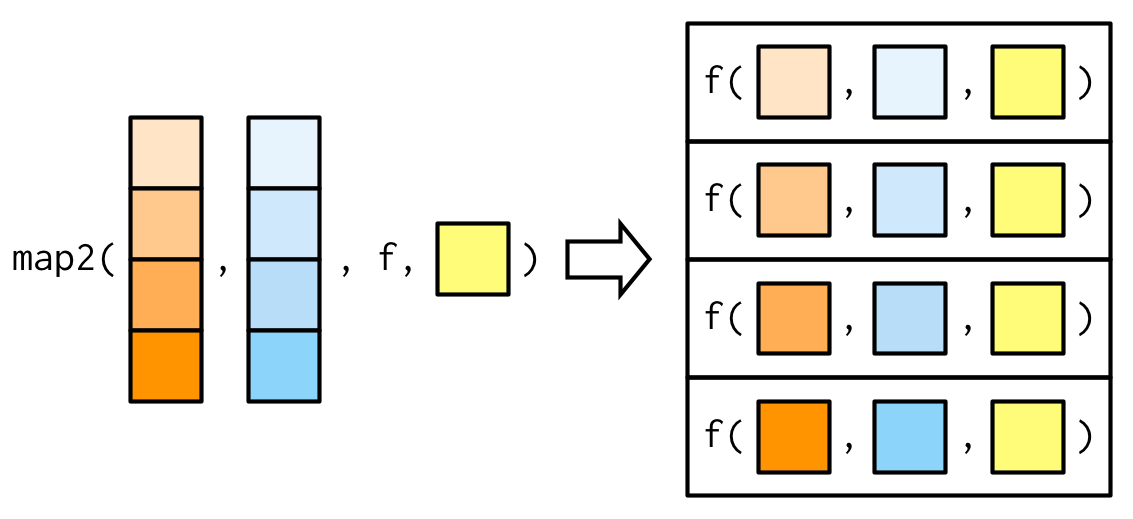
Source: Wickham (2019)
# passing na.rm = TRUEmap2_dbl(xs, ws, weighted.mean, na.rm = TRUE)## [1] 0.7355541 0.6625391 0.5968213 0.5287878purrr::map2()
Note that map2() also recycles inputs to ensure that they are the same length.
Example: weighted mean using map2() — ctd.
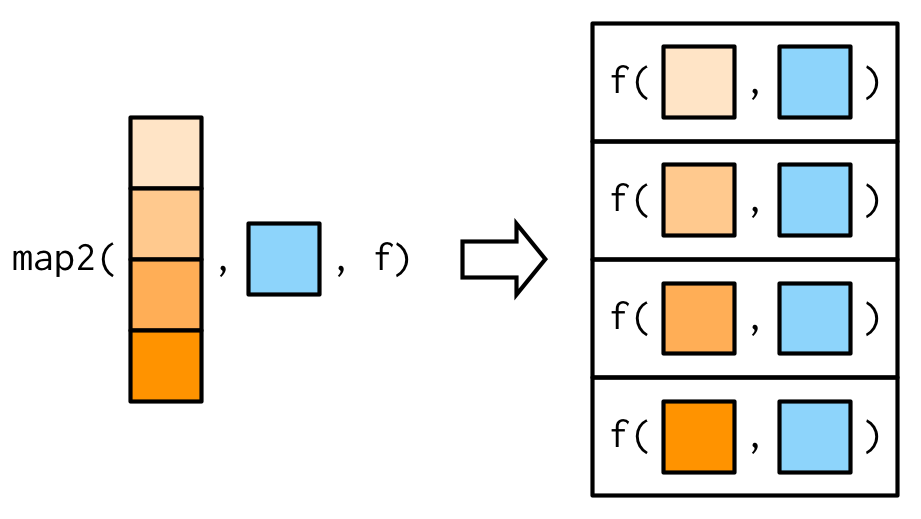
Source: Wickham (2019)
map2_dbl(1:6, 1, ~ .x + .y)## [1] 2 3 4 5 6 7purrr::walk()
walk()ignores the return value of.fand returns.xinvisibly. This is useful for functions that are called for their side-effects.There is no base R equivalent but wrapping
lapply()withinvisible()comes close
Example: assigning and passing objects
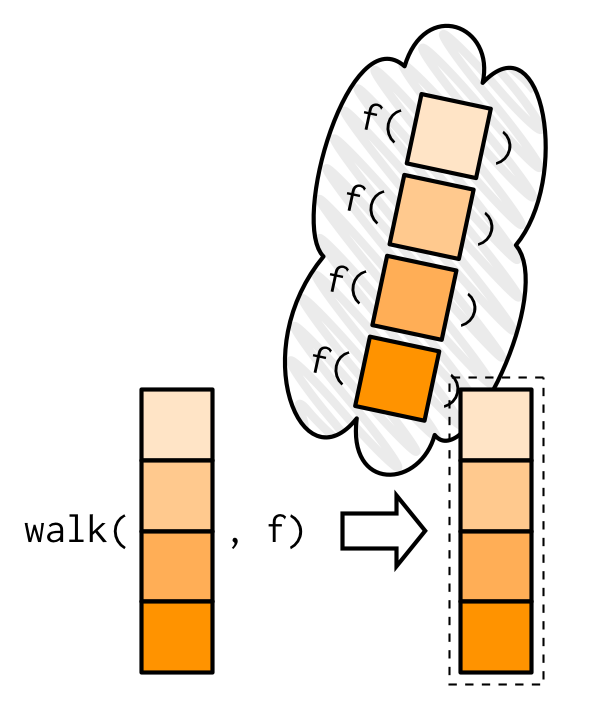
Source: Wickham (2019)
Assignment to an environment is a side-effect.

purrr::walk()
walk2() is a convenient alternative which is vectorised over two arguments.
Example: write to disc
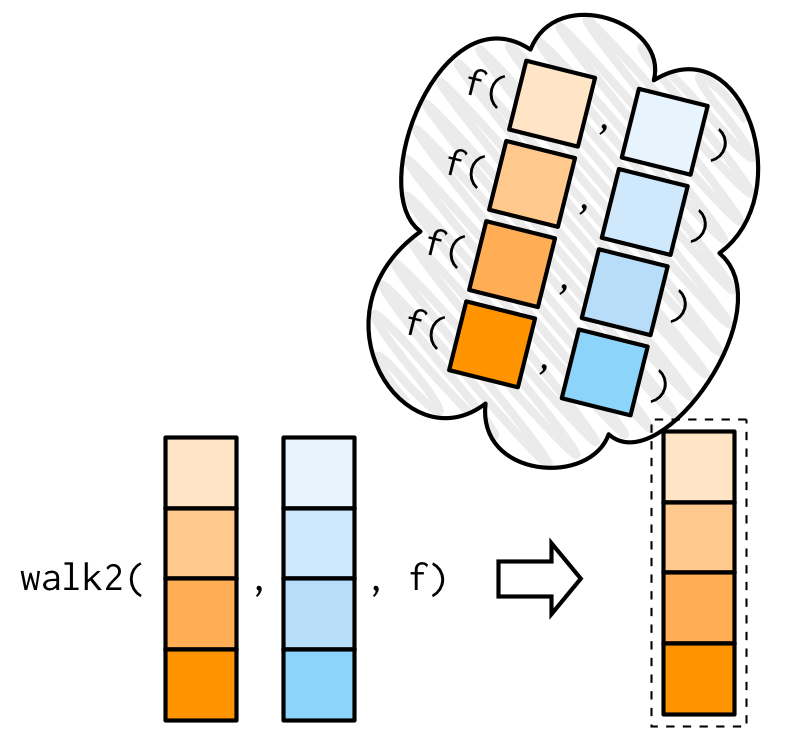
Source: Wickham (2019)
A common side-effect which needs two arguments (object and path) is writing to disk.

purrr::imap()
map(.x, .f)is essentially an analog tofor(x in xs) <apply .f to x and assign to list>for(i in seq_along(xs))andfor(nm in names(xs))are analogous toimap():imap(.x, .f)applies.fto values.xand indices or names derived from.x.
Example: named column means
imap() is a useful helper if we want to work with values along with variable names.

When using the formula shortcut, the first argument
.xis the value, and the second.yis the position/namecars2018 %>% select_if(is.numeric)returns a list so.yis a name and.xthe valuecharactervectors index by name,numericvectors index by position
purrr::pmap()
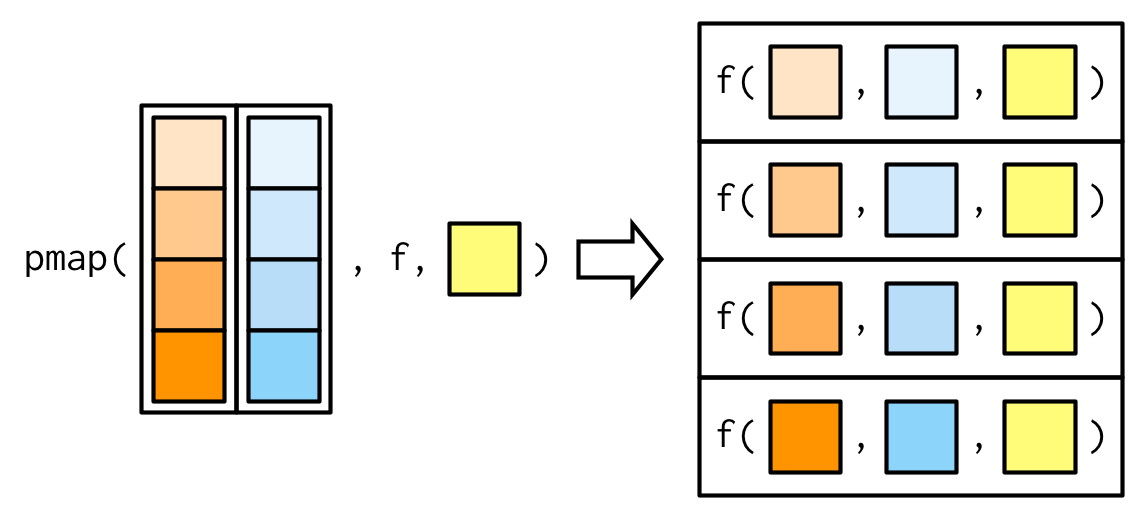
pmap() generalises map() and map2() to p vectorised arguments. Thus pmap(list(x, y), f) is the same as map2(x, y, f).
Example: weighted mean with pmap()
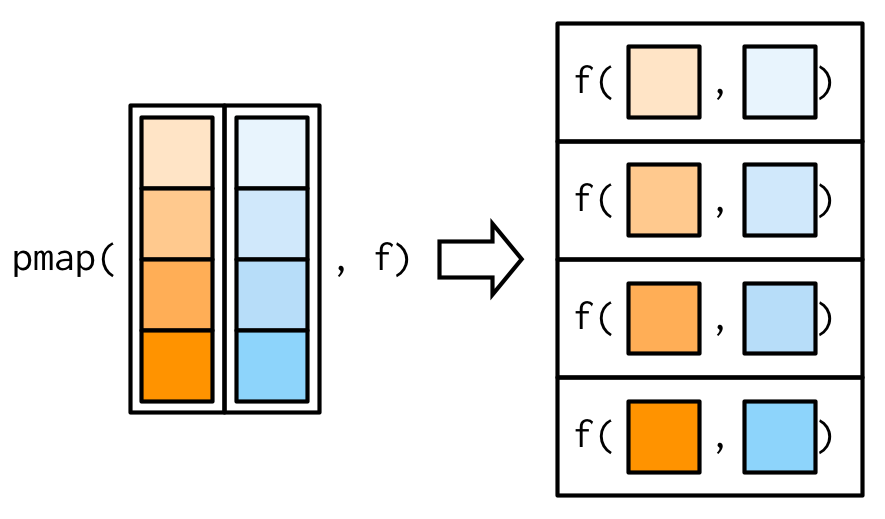
Source: Wickham (2019)
map2_dbl() behaves as pmap_dbl() in the two-argument case:
map2_dbl(xs, ws, weighted.mean)## [1] NA 0.6625391 0.5968213 0.5287878pmap_dbl(list(xs, ws), weighted.mean)## [1] NA 0.6625391 0.5968213 0.5287878purrr::pmap()
As before, additional arguments may be passed after .f and they are recycled, if necessary.
Example: weighted mean with pmap() — ctd.
Source: Wickham (2019)
Now with the additional argument na.rm = TRUE:
pmap_dbl(list(xs, ws), weighted.mean, na.rm = TRUE)## [1] 0.7355541 0.6625391 0.5968213 0.5287878purrr::pmap()
Note that pmap() gives much finer control over argument matching as we may use named list. This is very convenient for working with complex list objects.
Example: argument matching using named list

Source: Wickham (2019)
We look at the trimmed mean example again.
trims <- c(0, 0.1, 0.2, 0.5)x <- rcauchy(1000)Varying the trim argument can be done by passing the values in a named list.
pmap_dbl(list(trim = trims), mean, x = x)## [1] -0.03231537 0.06072652 0.04464511 0.04482772purrr::pmap()
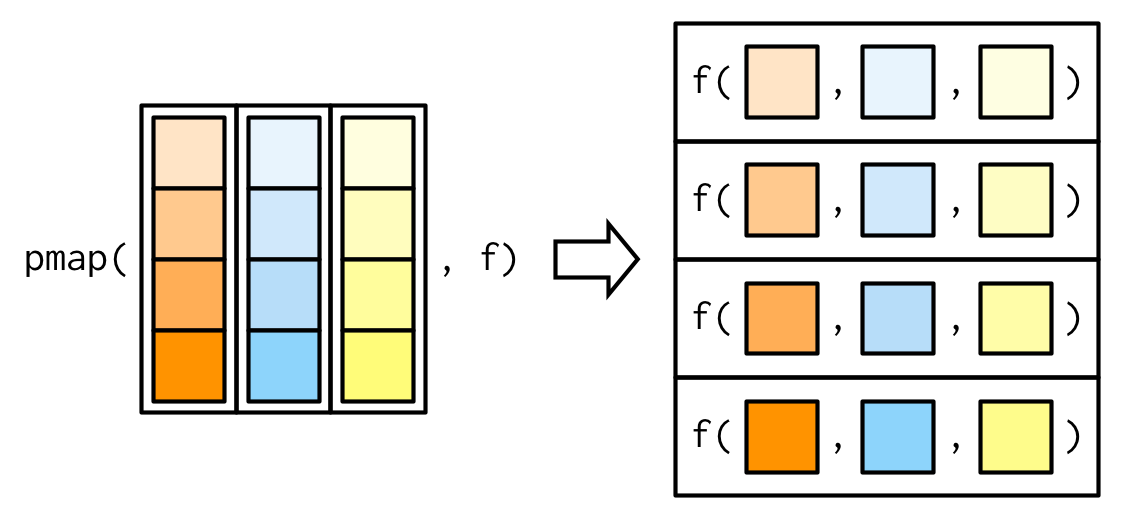
Remember that a data.frame is a list and thus can be passed to pmap() as a collection of inputs.
Example: pmap() with data.frame as input
Source: Wickham (2019)
params <- tibble::tribble( ~ n, ~ min, ~ max, 1L, 0, 1, 2L, 10, 100)Column names match the arguments: we don't have to worry about their order.
pmap(params, runif)## [[1]]## [1] 0.4475701## ## [[2]]## [1] 21.72593 65.95534tribble(): create tibbles using an easier to read row-by-row layout. This is useful for small tables of data where readability is important.
purrr::pmap() — Exercises
Explain the results of
modify(cars2018, 1)Explain how the following code transforms a
data.frameusing functions stored in alist.trans <- list(Displacement = function(x) x * 0.0163871,Transmission = function(x) factor(x, labels = c("Automatic", "Manual", "CVT")))nm <- names(trans)cars2018[nm] <- map2(trans, cars2018[nm], function(f, var) f(var))Compare and contrast the
map2()approach to thismap()approach:cars2018[nm] <- map(nm, ~ trans[[.x]](cars2018[[.x]]))
modify()is a shortcut forx[[i]] <- f(x[[i]]); return(x). So every row is filled with it's first value.Too lenghty, see class notes
As above
Part II
Function Factories
Function Factories
A function factory is a function that produces functions.
Example: function factory
nth_root <- function(n) { # function factory function(x) { x^(1/n) }}cube_root <- nth_root(3) # manufactured functioncube_root(8)## [1] 2Function Factories
The enclosing environment of the manufactured function is an execution environment of the function factory.
Example: function factory — ctd.
rlang::env_print(cube_root) # inspect enclosing environment## <environment: 0x1043a9fa8>## Parent: <environment: global>## Bindings:## • n: <dbl>rlang::fn_env(cube_root)$n # retrieve from enclosing environment## [1] 3Remember that execution environments are ephemeral in general: they are destroyed once the function has run.
This is different here: the enclosing environment of
cube_root()was the execution environment ofnth_root()—a mechanism which makes function factories possible.
Function Factories
Remember lazy evaluation?
Example: function factory — ctd.
n <- 2sq <- nth_root(n)n <- 16sq(64) # Wait... this should evaluate to 8!## [1] 1.2968464^(1/16) = 1.29684xis lazily evaluated whensq()is run, not whennth_root()is run. We thus need to force evaluation.This is likely to happen so it's a good practice to avoid such a bug by using
force()in your factories!
Function Factories
Remember lazy evaluation?
Example: function factory — ctd.
nth_root <- function(n) { force(n) function(x) { x^(1/n) }}n <- 2sq <- nth_root(n)n <- 16sq(64) # better :-)## [1] 8Question to students: why
force(x)and not justx? (check def. offorce()Note on Garbage Collection:
As manufactured functions hold on to the execution environment of the function factory you need to remove large objects manually.
f1 <- function(n) {x <- runif(n)m <- mean(x)rm(x) # use lobstr::obj_size() on a man. function to see differencefunction() m}
Function Factories — Stateful Functions
Function factories allow us to create functions with a memory.
Example: counter
# factory for a counternew_counter <- function() { i <- 0 function() { i <<- i + 1 i }}counter_one <- new_counter()counter_two <- new_counter()replicate(2, counter_one())## [1] 1 2replicate(5, counter_two())## [1] 1 2 3 4 5Should be used with moderation. The S6 system is more suitable if your manufactured functions are to manage multiple variables.
The "state" (
ihere) is tracked in the execution environment. This is a different environment for each function produced by the factory.rlang::env_print(counter_one)rlang::env_print(counter_two)
